PNU-159682CAS# 202350-68-3 |
- PF-562271
Catalog No.:BCC3674
CAS No.:717907-75-0
- TAE226 (NVP-TAE226)
Catalog No.:BCC3885
CAS No.:761437-28-9
- PF-573228
Catalog No.:BCC4496
CAS No.:869288-64-2
- PF-00562271
Catalog No.:BCC3684
CAS No.:939791-38-5
Quality Control & MSDS
Number of papers citing our products
Chemical structure

3D structure
| Cas No. | 202350-68-3 | SDF | Download SDF |
| PubChem ID | 9874188 | Appearance | Powder |
| Formula | C32H35NO13 | M.Wt | 641.62 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | DMSO : ≥ 100 mg/mL (155.86 mM) H2O : < 0.1 mg/mL (insoluble) *"≥" means soluble, but saturation unknown. | ||
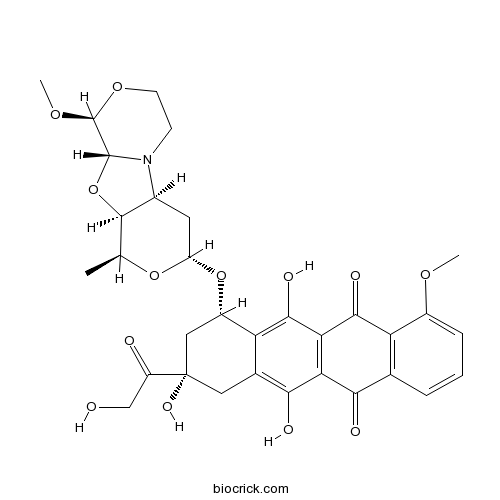
| Chemical Name | (7S,9S)-7-[[(1S,3R,4aS,9S,9aR,10aS)-9-methoxy-1-methyl-3,4,4a,6,7,9,9a,10a-octahydro-1H-pyrano[1,2][1,3]oxazolo[3,4-b][1,4]oxazin-3-yl]oxy]-6,9,11-trihydroxy-9-(2-hydroxyacetyl)-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione | ||
| SMILES | CC1C2C(CC(O1)OC3CC(CC4=C(C5=C(C(=C34)O)C(=O)C6=C(C5=O)C=CC=C6OC)O)(C(=O)CO)O)N7CCOC(C7O2)OC | ||
| Standard InChIKey | SLURUCSFDHKXFR-WWMWMSKMSA-N | ||
| Standard InChI | InChI=1S/C32H35NO13/c1-13-29-16(33-7-8-43-31(42-3)30(33)46-29)9-20(44-13)45-18-11-32(40,19(35)12-34)10-15-22(18)28(39)24-23(26(15)37)25(36)14-5-4-6-17(41-2)21(14)27(24)38/h4-6,13,16,18,20,29-31,34,37,39-40H,7-12H2,1-3H3/t13-,16-,18-,20-,29+,30+,31-,32-/m0/s1 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | PNU-159682, a highly potent metabolite of the anthracycline nemorubicin with outstanding cytotoxicity, is a potent ADCs cytotoxin.In Vitro:PNU-159682 inhibits a panel of human tumor cell lines with IC70 values in the range of 0.07-0.58 nM, and is 2,360- to 790-fold and 6,420- to 2,100-fold more potent than MMDX and doxorubicin, respectively[1]. PNU-159682 (100 μM) weakly inhibits topoisomerase II unknotting activity. PNU-159682 (10 μM)-DNA adducts contain one or two drug molecules bound to double-stranded DNA[2]. PNU-159682 shows cytotoxic effect on CAIX-expressing SKRC-52 cells with IC50 of 25 nM[3].In Vivo:PNU-159682 (15 μg/kg, i.v.) shows antitumor activity in mice bearing disseminated murine L1210 leukemia and in MX-1 human mammary carcinoma xenografts at 4 μg/kg[1]. PNU-159682 (25 nmol/kg) exhibits a potent antitumor effect in mice bearing SKRC-52 xenografted tumors[3]. References: | |||||

PNU-159682 Dilution Calculator

PNU-159682 Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 1.5586 mL | 7.7928 mL | 15.5855 mL | 31.1711 mL | 38.9639 mL |
| 5 mM | 0.3117 mL | 1.5586 mL | 3.1171 mL | 6.2342 mL | 7.7928 mL |
| 10 mM | 0.1559 mL | 0.7793 mL | 1.5586 mL | 3.1171 mL | 3.8964 mL |
| 50 mM | 0.0312 mL | 0.1559 mL | 0.3117 mL | 0.6234 mL | 0.7793 mL |
| 100 mM | 0.0156 mL | 0.0779 mL | 0.1559 mL | 0.3117 mL | 0.3896 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
PNU-159682 is a major bioactive metabolite of Nemorubicin in human liver microsomes; > 3,000-fold cytotoxic than its parent compound(MMDX and doxorubicin).
- Dasycarpol
Catalog No.:BCN7134
CAS No.:202343-57-5
- Dregeoside Aa1
Catalog No.:BCN4678
CAS No.:20230-41-5
- Spiraeoside
Catalog No.:BCC8251
CAS No.:20229-56-5
- Flucytosine
Catalog No.:BCC3780
CAS No.:2022-85-7
- Spiperone hydrochloride
Catalog No.:BCC6882
CAS No.:2022-29-9
- Bilastine
Catalog No.:BCC5263
CAS No.:202189-78-4
- Tenofovir Disoproxil Fumarate
Catalog No.:BCC1108
CAS No.:202138-50-9
- Kaempferol 7-O-rhamnoside
Catalog No.:BCN6489
CAS No.:20196-89-8
- LY341495
Catalog No.:BCC1724
CAS No.:201943-63-7
- Epicatechin pentaacetate
Catalog No.:BCN4884
CAS No.:20194-41-6
- PTAC oxalate
Catalog No.:BCC6217
CAS No.:201939-40-4
- Ombuoside
Catalog No.:BCN3711
CAS No.:20188-85-6
- Scillascillol
Catalog No.:BCN7008
CAS No.:2023822-39-9
- Scillascillone
Catalog No.:BCN6992
CAS No.:2023822-40-2
- Scillascilloside B-1
Catalog No.:BCN6998
CAS No.:2023822-41-3
- 2,2-Bis(4-chloroformyloxyphenyl)propane
Catalog No.:BCC8491
CAS No.:2024-88-6
- Etoricoxib
Catalog No.:BCC1565
CAS No.:202409-33-4
- Hydroxygenkwanin
Catalog No.:BCN4885
CAS No.:20243-59-8
- 1,3,7-Trihydroxy-2-prenylxanthone
Catalog No.:BCN4886
CAS No.:20245-39-0
- AM 281
Catalog No.:BCC6944
CAS No.:202463-68-1
- Ro 04-6790
Catalog No.:BCC7512
CAS No.:202466-68-0
- Glomeratose A
Catalog No.:BCN8400
CAS No.:202471-84-9
- JANEX-1
Catalog No.:BCC1668
CAS No.:202475-60-3
- 13(18)-Oleanen-3-one
Catalog No.:BCN4887
CAS No.:20248-08-2
The interaction of nemorubicin metabolite PNU-159682 with DNA fragments d(CGTACG)(2), d(CGATCG)(2) and d(CGCGCG)(2) shows a strong but reversible binding to G:C base pairs.[Pubmed:23154079]
Bioorg Med Chem. 2012 Dec 15;20(24):6979-88.
The antitumor anthracycline nemorubicin is converted by human liver microsomes to a major metabolite, PNU-159682 (PNU), which was found to be much more potent than its parent drug toward cultured tumor cells and in vivo tumor models. The mechanism of action of nemorubicin appears different from other anthracyclines and until now is the object of studies. In fact PNU is deemed to play a dominant, but still unclear, role in the in vivo antitumor activity of nemorubicin. The interaction of PNU with the oligonucleotides d(CGTACG)(2), d(CGATCG)(2) and d(CGCGCG)(2) was studied with a combined use of (1)H and (31)P NMR spectroscopy and by ESI-mass experiments. The NMR studies allowed to establish that the intercalation between the base pairs of the duplex leads to very stable complexes and at the same time to exclude the formation of covalent bonds. Melting experiments monitored by NMR, allowed to observe with high accuracy the behaviour of the imine protons with temperature, and the results showed that the re-annealing occurs after melting. The formation of reversible complexes was confirmed by HPLC-tandem mass spectra, also combined with endonuclease P1digestion. The MS/MS spectra showed the loss of neutral PNU before breaking the double helix, a behaviour typical of intercalators. After digestion with the enzyme, the spectra did not show any compound with PNU bound to the bases. The evidence of a reversible process appears from both proton and phosphorus NOESY spectra of PNU bound to d(CGTACG)(2) and to d(CGATCG)(2). The dissociation rate constants (k(off)) of the slow step of the intercalation process, measured by (31)P NMR NOE-exchange experiments, showed that the kinetics of the process is slower for PNU than for doxorubicin and nemorubicin, leading to a 10- to 20-fold increase of the residence time of PNU into the intercalation sites, with respect to doxorubicin. A relevant number of NOE interactions allowed to derive a model of the complexes in solution from restrained MD calculations. The conformation of PNU bound to the oligonucleotides was also derived from the coupling constant values.
Virtual Cross-Linking of the Active Nemorubicin Metabolite PNU-159682 to Double-Stranded DNA.[Pubmed:28068470]
Chem Res Toxicol. 2017 Feb 20;30(2):614-624.
The DNA alkylating mechanism of PNU-159682 (PNU), a highly potent metabolite of the anthracycline nemorubicin, was investigated by gel-electrophoretic, HPLC-UV, and micro-HPLC/mass spectrometry (MS) measurements. PNU quickly reacted with double-stranded oligonucleotides, but not with single-stranded sequences, to form covalent adducts which were detectable by denaturing polyacrylamide gel electrophoresis (DPAGE). Ion-pair reverse-phase HPLC-UV analysis on CG rich duplex sequences having a 5'-CCCGGG-3' central core showed the formation of two types of adducts with PNU, which were stable and could be characterized by micro-HPLC/MS. The first type contained one alkylated species (and possibly one reversibly bound species), and the second contained two alkylated species per duplex DNA. The covalent adducts were found to produce effective bridging of DNA complementary strands through the formation of virtual cross-links reminiscent of those produced by classical anthracyclines in the presence of formaldehyde. Furthermore, the absence of reactivity of PNU with CG-rich sequence containing a TA core (CGTACG), and the minor reactivity between PNU and CGC sequences (TACGCG.CGCGTA) pointed out the importance of guanine sequence context in modulating DNA alkylation.
Formation and antitumor activity of PNU-159682, a major metabolite of nemorubicin in human liver microsomes.[Pubmed:15746066]
Clin Cancer Res. 2005 Feb 15;11(4):1608-17.
PURPOSE: Nemorubicin (3'-deamino-3'-[2''(S)-methoxy-4''-morpholinyl]doxorubicin; MMDX) is an investigational drug currently in phase II/III clinical testing in hepatocellular carcinoma. A bioactivation product of MMDX, 3'-deamino-3'',4'-anhydro-[2''(S)-methoxy-3''(R)-oxy-4''-morpholinyl]doxorubicin (PNU-159682), has been recently identified in an incubate of the drug with NADPH-supplemented rat liver microsomes. The aims of this study were to obtain information about MMDX biotransformation to PNU-159682 in humans, and to explore the antitumor activity of PNU-159682. EXPERIMENTAL DESIGN: Human liver microsomes (HLM) and microsomes from genetically engineered cell lines expressing individual human cytochrome P450s (CYP) were used to study MMDX biotransformation. We also examined the cytotoxicity and antitumor activity of PNU-159682 using a panel of in vitro-cultured human tumor cell lines and tumor-bearing mice, respectively. RESULTS: HLMs converted MMDX to a major metabolite, whose retention time in liquid chromatography and ion fragmentation in tandem mass spectrometry were identical to those of synthetic PNU-159682. In a bank of HLMs from 10 donors, rates of PNU-159682 formation correlated significantly with three distinct CYP3A-mediated activities. Troleandomycin and ketoconazole, both inhibitors of CYP3A, markedly reduced PNU-159682 formation by HLMs; the reaction was also concentration-dependently inhibited by a monoclonal antibody to CYP3A4/5. Of the 10 cDNA-expressed CYPs examined, only CYP3A4 formed PNU-159682. In addition, PNU-159682 was remarkably more cytotoxic than MMDX and doxorubicin in vitro, and was effective in the two in vivo tumor models tested, i.e., disseminated murine L1210 leukemia and MX-1 human mammary carcinoma xenografts. CONCLUSIONS: CYP3A4, the major CYP in human liver, converts MMDX to a more cytotoxic metabolite, PNU-159682, which retains antitumor activity in vivo.
In vitro hepatic conversion of the anticancer agent nemorubicin to its active metabolite PNU-159682 in mice, rats and dogs: a comparison with human liver microsomes.[Pubmed:18671948]
Biochem Pharmacol. 2008 Sep 15;76(6):784-95.
We recently demonstrated that nemorubicin (MMDX), an investigational antitumor drug, is converted to an active metabolite, PNU-159682, by human liver cytochrome P450 (CYP) 3A4. The objectives of this study were: (1) to investigate MMDX metabolism by liver microsomes from laboratory animals (mice, rats, and dogs of both sexes) to ascertain whether PNU-159682 is also produced in these species, and to identify the CYP form(s) responsible for its formation; (2) to compare the animal metabolism of MMDX with that by human liver microsomes (HLMs), in order to determine which animal species is closest to human beings; (3) to explore whether differences in PNU-159682 formation are responsible for previously reported species- and sex-related differences in MMDX host toxicity. The animal metabolism of MMDX proved to be qualitatively similar to that observed with HLMs since, in all tested species, MMDX was mainly converted to PNU-159682 by a single CYP3A form. However, there were marked quantitative inter- and intra-species differences in kinetic parameters. The mouse and the male rat exhibited V(max) and intrinsic metabolic clearance (CL(int)) values closest to those of human beings, suggesting that these species are the most suitable animal models to investigate MMDX biotransformation. A close inverse correlation was found between MMDX CL(int) and previously reported values of MMDX LD(50) for animals of the species, sex and strain tested here, indicating that differences in the in vivo toxicity of MMDX are most probably due to sex- and species-related differences in the extent of PNU-159682 formation.


