P 22077USP7/(DUB)USP47 inhibitor CAS# 1247819-59-5 |
- USP7-USP47 inhibitor
Catalog No.:BCC4113
CAS No.:1247825-37-1
- NSC 632839 hydrochloride
Catalog No.:BCC2088
CAS No.:157654-67-6
- PR-619
Catalog No.:BCC3627
CAS No.:2645-32-1
- WP1130
Catalog No.:BCC3686
CAS No.:856243-80-6
- Vialinin A
Catalog No.:BCC2367
CAS No.:858134-23-3
- P005091
Catalog No.:BCC1287
CAS No.:882257-11-6
Quality Control & MSDS
Number of papers citing our products
Chemical structure
3D structure
| Cas No. | 1247819-59-5 | SDF | Download SDF |
| PubChem ID | 46931953 | Appearance | Powder |
| Formula | C12H7F2NO3S2 | M.Wt | 315.32 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | DMSO : 50 mg/mL (158.57 mM; Need ultrasonic) | ||
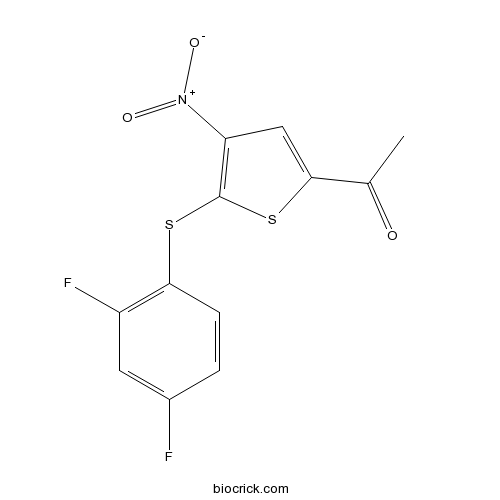
| Chemical Name | 1-[5-(2,4-difluorophenyl)sulfanyl-4-nitrothiophen-2-yl]ethanone | ||
| SMILES | CC(=O)C1=CC(=C(S1)SC2=C(C=C(C=C2)F)F)[N+](=O)[O-] | ||
| Standard InChIKey | RMAMGGNACJHXHO-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C12H7F2NO3S2/c1-6(16)11-5-9(15(17)18)12(20-11)19-10-3-2-7(13)4-8(10)14/h2-5H,1H3 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | Inhibitor of ubiquitin-specific protease (USP) 7 (EC50 = 8.6 μM); also inhibits the closely related deubiquitinase (DUB) USP47. Demonstrates downstream inhibition of HDM2 and claspin in vitro. Inhibits USP7-mediated p53 deubiquitination. Blocks deubiquitination of Tip60 (KAT5) histone lysine acetyltransferase. |

P 22077 Dilution Calculator

P 22077 Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 3.1714 mL | 15.8569 mL | 31.7138 mL | 63.4276 mL | 79.2845 mL |
| 5 mM | 0.6343 mL | 3.1714 mL | 6.3428 mL | 12.6855 mL | 15.8569 mL |
| 10 mM | 0.3171 mL | 1.5857 mL | 3.1714 mL | 6.3428 mL | 7.9285 mL |
| 50 mM | 0.0634 mL | 0.3171 mL | 0.6343 mL | 1.2686 mL | 1.5857 mL |
| 100 mM | 0.0317 mL | 0.1586 mL | 0.3171 mL | 0.6343 mL | 0.7928 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
Description:IC50: 8.6 nM (singleplexed); 7.8 nM (multiplexed)
USP7, also known as HAUSP, has been found to be critical in cancer progression due to its influence on the stability of the tumor suppressor p53. P022077 Inhibits ubiquitin-specific protease (USP) 7 and also inhibits the closely related deubiquitinase (DUB) USP4.
In vitro: P022077 had negligible activity versus DEN1 and SENP2core over the concentration range tested, but inhibited USP7 with an IC50 of 8 μM [1]. In another study Inhibiting, USP7 with the small-molecule inhibitor P22077 attenuates the p53-dependent apoptotic pathway by destabilizing Tip60. P22077, however, is still cytotoxic, and this is partly due to destabilization of Tip60 [2].
In vivo: P022077 is currently in in-vitro investigation and no animal in vivo study is ongoing.
Clinical trial: P022077 is currently in the preclinical development and no clinical trial is ongoing.
References:
[1] Tian X, Isamiddinova NS, Peroutka RJ, Goldenberg SJ, Mattern MR, Nicholson B, Leach C. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay Drug Dev Technol. 2011;9(2):165-73.
[2] Dar A, Shibata E, Dutta A. Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Mol Cell Biol. 2013;33(16):3309-20.
- Losartan Potassium (DuP 753)
Catalog No.:BCC1080
CAS No.:124750-99-8
- Quinovic acid 3-O-(6-deoxy-beta-D-glucopyranoside) 28-O-beta-D-glucopyranosyl ester
Catalog No.:BCN1595
CAS No.:124727-10-2
- Meprednisone
Catalog No.:BCC4893
CAS No.:1247-42-3
- Methyl-Dodovisate A
Catalog No.:BCN4719
CAS No.:1246937-33-6
- 5'-Prenylaliarin
Catalog No.:BCN4829
CAS No.:1246926-09-9
- 5,7,4'-Trihydroxy-3,6-dimethoxy-3',5'-diprenylflavone
Catalog No.:BCN1596
CAS No.:1246926-08-8
- VS-5584 (SB2343)
Catalog No.:BCC2047
CAS No.:1246560-33-7
- MPI-0479605
Catalog No.:BCC5347
CAS No.:1246529-32-7
- SR1078
Catalog No.:BCC1963
CAS No.:1246525-60-9
- (-)-Dihydroguaiaretic acid
Catalog No.:BCN8002
CAS No.:124649-78-1
- Officinaruminane B
Catalog No.:BCN3594
CAS No.:1246282-67-6
- C3bot (154-182)
Catalog No.:BCC6117
CAS No.:1246280-79-4
- USP7-USP47 inhibitor
Catalog No.:BCC4113
CAS No.:1247825-37-1
- Valaciclovir
Catalog No.:BCC2025
CAS No.:124832-26-4
- Valacyclovir hydrochloride
Catalog No.:BCC4051
CAS No.:124832-27-5
- Nadifloxacin
Catalog No.:BCC4804
CAS No.:124858-35-1
- 5-Dehydroxyparatocarpin K
Catalog No.:BCN4017
CAS No.:124858-37-3
- Isoaltholactone
Catalog No.:BCN4826
CAS No.:124868-11-7
- Benzoyl leuco methylene blue
Catalog No.:BCC8862
CAS No.:1249-97-4
- (R)-(+)-Tolterodine
Catalog No.:BCC4054
CAS No.:124937-51-5
- Tolterodine tartrate
Catalog No.:BCC4586
CAS No.:124937-52-6
- Tectoroside
Catalog No.:BCN6672
CAS No.:124960-89-0
- Raddeanoside R8
Catalog No.:BCN6555
CAS No.:124961-61-1
- Ajugacumbin A
Catalog No.:BCN3657
CAS No.:124961-66-6
Phenotypic spectrum of Costello syndrome individuals harboring the rare HRAS mutation p.Gly13Asp.[Pubmed:28371260]
Am J Med Genet A. 2017 May;173(5):1309-1318.
Costello syndrome is part of the RASopathies, a group of neurocardiofaciocutaneous syndromes caused by deregulation of the RAS mitogen-activated protein kinase pathway. Heterozygous mutations in HRAS are responsible for Costello syndrome, with more than 80% of the patients harboring the specific p.Gly12Ser variant. These individuals show a homogeneous phenotype. The clinical characteristics of the Costello syndrome individuals harboring rarer HRAS mutations are less understood, due to the small number of reported cases. Here, we describe the phenotypic spectrum of five additional individuals with HRAS c.38G>A; p.Gly13Asp, including one with somatic mosaicism, and review five previously described cases. The facial and hair abnormalities of the HRAS p.Gly13Asp individuals differ from the typical pattern observed in those showing the common HRAS (p.Gly12Ser) mutation, with less coarse facial features and slow growing, sparse hair with abnormal texture, the latter resembling the pattern observed in Noonan syndrome-like disorder with loose anagen hair and individuals harboring another amino acid substitution in HRAS (p.Gly13Cys). Although some individuals with HRAS p.Gly13Asp developed papillomata and vascular proliferation lesions, no malignant tumors occurred, similar to what was reported for individuals harboring the HRAS p.Gly13Cys. The fact that no malignant tumors were described in these individuals does not allow definitive conclusions about the risk for cancer development. It remains to be determined if substitutions of amino acid 13 in HRAS (p.Gly13Asp and p.Gly13Cys) increase the risk of tumor development.
Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.[Pubmed:28370477]
Mol Microbiol. 2017 Jul;105(1):25-45.
Catabolite repression is a mechanism that enables bacteria to control carbon utilization. As part of this global regulatory network, components of the phosphoenolpyruvate:carbohydrate phosphotransferase system inhibit the uptake of less favorable sugars when a preferred carbon source such as glucose is available. This process is termed inducer exclusion. In bacteria belonging to the phylum Firmicutes, HPr, phosphorylated at serine 46 (P-Ser46-HPr) is the key player but its mode of action is elusive. To address this question at the level of purified protein components, we have chosen a homolog of the Escherichia coli maltose/maltodextrin ATP-binding cassette transporter from Lactobacillus casei (MalE1-MalF1G1K12 ) as a model system. We show that the solute binding protein, MalE1, binds linear and cyclic maltodextrins but not maltose. Crystal structures of MalE1 complexed with these sugars provide a clue why maltose is not a substrate. P-Ser46-HPr inhibited MalE1/maltotetraose-stimulated ATPase activity of the transporter incorporated in proteoliposomes. Furthermore, cross-linking experiments revealed that P-Ser46-HPr contacts the nucleotide-binding subunit, MalK1, in proximity to the Walker A motif. However, P-Ser46-HPr did not block binding of ATP to MalK1. Together, our findings provide first biochemical evidence that P-Ser-HPr arrests the transport cycle by preventing ATP hydrolysis at the MalK1 subunits of the transporter.
The Lightest Element Phosphoranylidene: NHC-Supported Cyclic Borylidene-Phosphorane with Significant B=P Character.[Pubmed:28371066]
Angew Chem Int Ed Engl. 2017 Apr 18;56(17):4814-4818.
A borylidene-phosphorane, the lightest phosphoranylidene, which is stabilized by an N-heterocyclic carbene ligand, was synthesized and fully characterized. Experimental electron density analysis and DFT calculations indicate an enhanced ylene character (rather than an ylide character) with an exceptionally strong B-->P pi-back bonding related to the less electronegative boron compared to phosphorus.
PCA Based on Graph Laplacian Regularization and P-Norm for Gene Selection and Clustering.[Pubmed:28371780]
IEEE Trans Nanobioscience. 2017 Jun;16(4):257-265.
In modern molecular biology, the hotspots and difficulties of this field are identifying characteristic genes from gene expression data. Traditional reconstruction-error-minimization model principal component analysis (PCA) as a matrix decomposition method uses quadratic error function, which is known sensitive to outliers and noise. Hence, it is necessary to learn a good PCA method when outliers and noise exist. In this paper, we develop a novel PCA method enforcing P-norm on error function and graph-Laplacian regularization term for matrix decomposition problem, which is called as PgLPCA. The heart of the method designing for reducing outliers and noise is a new error function based on non-convex proximal P-norm. Besides, Laplacian regularization term is used to find the internal geometric structure in the data representation. To solve the minimization problem, we develop an efficient optimization algorithm based on the augmented Lagrange multiplier method. This method is used to select characteristic genes and cluster the samples from explosive biological data, which has higher accuracy than compared methods.
Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway.[Pubmed:23775119]
Mol Cell Biol. 2013 Aug;33(16):3309-20.
Tip60 is an essential acetyltransferase required for acetylation of nucleosomal histones and other nonhistone proteins. Tip60 acetylates the p53 tumor suppressor at lysine 120 (K120), a modification essential for p53-dependent induction of PUMA and apoptosis. It is known that Tip60 is turned over in cells by the ubiquitin-proteasome system. However, the deubiquitinase activity for stabilizing Tip60 is unknown. Here we show that USP7 interacts with and deubiquitinates Tip60 both in vitro and in vivo. USP7 deubiquitinase activity is required for the stabilization of Tip60 in order to operate an effective p53-dependent apoptotic pathway in response to genotoxic stress. Inhibiting USP7 with the small-molecule inhibitor P22077 attenuates the p53-dependent apoptotic pathway by destabilizing Tip60. P22077, however, is still cytotoxic, and this is partly due to destabilization of Tip60.
Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes.[Pubmed:22118674]
Chem Biol. 2011 Nov 23;18(11):1401-12.
Converting lead compounds into drug candidates is a crucial step in drug development, requiring early assessment of potency, selectivity, and off-target effects. We have utilized activity-based chemical proteomics to determine the potency and selectivity of deubiquitylating enzyme (DUB) inhibitors in cell culture models. Importantly, we characterized the small molecule PR-619 as a broad-range DUB inhibitor, and P22077 as a USP7 inhibitor with potential for further development as a chemotherapeutic agent in cancer therapy. A striking accumulation of polyubiquitylated proteins was observed after both selective and general inhibition of cellular DUB activity without direct impairment of proteasomal proteolysis. The repertoire of ubiquitylated substrates was analyzed by tandem mass spectrometry, identifying distinct subsets for general or specific inhibition of DUBs. This enabled identification of previously unknown functional links between USP7 and enzymes involved in DNA repair.
Deubiquitylase, deSUMOylase, and deISGylase activity microarrays for assay of substrate preference and functional modifiers.[Pubmed:20956615]
Mol Cell Proteomics. 2011 Jan;10(1):M110.002402.
Microarray-based proteomics expanded the information potential of DNA arrays to the level of protein translation and interaction, but so far, not much beyond. Although enzymatic activity from immobilized proteins has been reliably studied using surface plasmon resonance, a microarray of catalytically competent enzymes would facilitate high throughput, parallel study of their function. The ability to localize activity from soluble substrates has frustrated development of such an array. Here, we report the novel use of previously developed, highly specific suicide substrates for three families of enzymes: deubiquitylases, deSUMOylases, and deISGylases. We show specificity of each family to its cognate substrate, and demonstrate utility of the array in a secondary screen of small molecule inhibitors.
Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format.[Pubmed:21133675]
Assay Drug Dev Technol. 2011 Apr;9(2):165-73.
The reversible conjugation of ubiquitin and ubiquitin-like (UbL) proteins to protein substrates plays a critical role in the regulation of many cellular pathways. The removal of ubiquitin from target proteins is performed by ubiquitin proteases also known as deubiquitylases (DUBs). Owing to their substrate specificity and the central role ubiquitylation plays in cell signaling pathways, DUB are attractive targets for therapeutic development. The development of DUB inhibitors requires assays that are amenable to high-throughput screening and provide rapid assessment of inhibitor selectivity. Determination of inhibitor selectivity at an early stage of drug discovery will reduce drug failure in the clinic as well as reduce overall drug development costs. We have developed two novel assays, UbL-Enterokinase light chain and UbL-Granzyme B, for quantifying ubiquitin and UbL protease activity. In our quest to discover and characterize novel chemical entities, we have combined these assays with a previously developed assay in a multiplex format. This multiplex format allows for the detection of three distinct protease activities simultaneously, in a single well. We have demonstrated that the multiplex format is able to distinguish between selective and nonselective protease inhibitors. Specifically, we have used this assay format to characterize P022077, a selective ubiquitin-specific protease 7 inhibitor discovered at Progenra.


