GuanineCAS# 73-40-5 |
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products
| Cas No. | 73-40-5 | SDF | Download SDF |
| PubChem ID | 764 | Appearance | Powder |
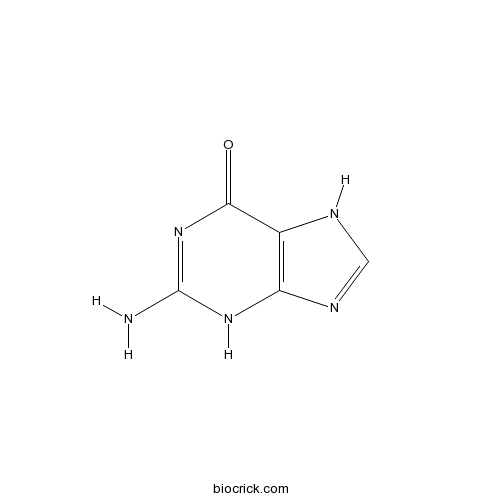
| Formula | C5H5N5O | M.Wt | 151.13 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | DMSO : 0.15 mg/mL (0.99 mM; Need ultrasonic and warming) | ||
| Chemical Name | 2-amino-3,7-dihydropurin-6-one | ||
| SMILES | C1=NC2=C(N1)C(=O)N=C(N2)N | ||
| Standard InChIKey | UYTPUPDQBNUYGX-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C5H5N5O/c6-5-9-3-2(4(11)10-5)7-1-8-3/h1H,(H4,6,7,8,9,10,11) | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Guanine Dilution Calculator

Guanine Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 6.6168 mL | 33.0841 mL | 66.1682 mL | 132.3364 mL | 165.4205 mL |
| 5 mM | 1.3234 mL | 6.6168 mL | 13.2336 mL | 26.4673 mL | 33.0841 mL |
| 10 mM | 0.6617 mL | 3.3084 mL | 6.6168 mL | 13.2336 mL | 16.542 mL |
| 50 mM | 0.1323 mL | 0.6617 mL | 1.3234 mL | 2.6467 mL | 3.3084 mL |
| 100 mM | 0.0662 mL | 0.3308 mL | 0.6617 mL | 1.3234 mL | 1.6542 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
- H-Ile-OH
Catalog No.:BCC2960
CAS No.:73-32-5
- Melatonin
Catalog No.:BCN2196
CAS No.:73-31-4
- Adenine
Catalog No.:BCC4450
CAS No.:73-24-5
- H-Trp-OH
Catalog No.:BCC3111
CAS No.:73-22-3
- Cordycepin
Catalog No.:BCN5389
CAS No.:73-03-0
- L-(-)-threo-3-Hydroxyaspartic acid
Catalog No.:BCC6565
CAS No.:7298-99-9
- Brassinolide
Catalog No.:BCC1438
CAS No.:72962-43-7
- BMS-707035
Catalog No.:BCC2133
CAS No.:729607-74-3
- XRP44X
Catalog No.:BCC7568
CAS No.:729605-21-4
- Gomisin O
Catalog No.:BCN2875
CAS No.:72960-22-6
- Gomisin E
Catalog No.:BCN7031
CAS No.:72960-21-5
- 6,7-Dihydroneridienone A
Catalog No.:BCN4020
CAS No.:72959-46-7
- Lidocaine hydrochloride
Catalog No.:BCC9009
CAS No.:73-78-9
- 15-Isopimarene-8,18-diol
Catalog No.:BCN4287
CAS No.:73002-86-5
- CyPPA
Catalog No.:BCC7526
CAS No.:73029-73-9
- Atractylenolide III
Catalog No.:BCN1045
CAS No.:73030-71-4
- Epigomisin O
Catalog No.:BCN2862
CAS No.:73036-31-4
- Atractylenolide I
Catalog No.:BCN1043
CAS No.:73069-13-3
- Atractylenolide II
Catalog No.:BCN1044
CAS No.:73069-14-4
- Praeruptorin A
Catalog No.:BCN4987
CAS No.:73069-25-7
- (+)-Praeruptorin A
Catalog No.:BCN4989
CAS No.:73069-27-9
- Praeruptorin D
Catalog No.:BCN4990
CAS No.:73069-28-0
- Scutebarbatine D
Catalog No.:BCN8536
CAS No.:910099-76-2
- Arecoline
Catalog No.:BCN8537
CAS No.:73069-28-9
Pathways of Metabolite-Related Damage to a Synthetic p53 Gene Exon 7 Oligonucleotide Using Magnetic Enzyme Bioreactor Beads and LC-MS/MS Sequencing.[Pubmed:29750510]
Biochemistry. 2018 Jul 3;57(26):3883-3893.
Reactive metabolites of environmental chemicals and drugs can cause site specific damage to the p53 tumor suppressor gene in a major pathway for genotoxicity. We report here a high-throughput, cell-free, 96-well plate magnetic bead-enzyme system interfaced with LC-MS/MS sequencing for bioactivating test chemicals and identifying resulting adduction sites on genes. Bioactivated aflatoxin B1 was reacted with a 32 bp exon 7 fragment of the p53 gene using eight microsomal cytochrome (cyt) P450 enzymes from different organs coated on magnetic beads. All cyt P450s converted aflatoxin B1 to aflatoxin B1-8,9-epoxide that adducts Guanine (G) in codon 249, with subsequent depurination to give abasic sites and then strand breaks. This is the first demonstration in a cell-free medium that the aflatoxin B1 metabolite selectively causes abasic site formation and strand breaks at codon 249 of the p53 probe, corresponding to the chemical pathway and mutations of p53 in human liver cells and tumors. Molecular modeling supports the view that binding of aflatoxin B1-8,9-epoxide to G in codon 249 precedes the SN2 adduction reaction. Among a range of metabolic enzymes characteristic of different organs, human liver microsomes and cyt P450 3A5 supersomes showed the highest bioactivation rate for p53 exon 7 damage. This method of identifying metabolite-related gene damage sites may facilitate predictions of organ specific cancers for test chemicals via correlations with mutation sites.
Targeting Photoinduced DNA Destruction by Ru(II) Tetraazaphenanthrene in Live Cells by Signal Peptide.[Pubmed:29767962]
J Am Chem Soc. 2018 Jun 6;140(22):6945-6955.
Exploiting NF-kappaB transcription factor peptide conjugation, a Ru(II)-bis-tap complex (tap = 1,4,5,8-tetraazaphenanthrene) was targeted specifically to the nuclei of live HeLa and CHO cells for the first time. DNA binding of the complex within the nucleus of live cells was evident from gradual extinction of the metal complex luminescence after it had crossed the nuclear envelope, attributed to Guanine quenching of the ruthenium emission via photoinduced electron transfer. Resonance Raman imaging confirmed that the complex remained in the nucleus after emission is extinguished. In the dark and under imaging conditions the cells remain viable, but efficient cellular destruction was induced with precise spatiotemporal control by applying higher irradiation intensities to selected cells. Solution studies indicate that the peptide conjugated complex associates strongly with calf thymus DNA ex-cellulo and gel electrophoresis confirmed that the peptide conjugate is capable of singlet oxygen independent photodamage to plasmid DNA. This indicates that the observed efficient cellular destruction likely operates via direct DNA oxidation by photoinduced electron transfer between Guanine and the precision targeted Ru(II)-tap probe. The discrete targeting of polyazaaromatic complexes to the cell nucleus and confirmation that they are photocytotoxic after nuclear delivery is an important step toward their application in cellular phototherapy.
Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods.[Pubmed:29765080]
Sci Rep. 2018 May 15;8(1):7560.
Mirtrons are non-canonical microRNAs encoded in introns the biogenesis of which starts with splicing. They are not processed by Drosha and enter the canonical pathway at the Exportin-5 level. Mirtrons are much less evolutionary conserved than canonical miRNAs. Due to the differences, canonical miRNA predictors are not applicable to mirtron prediction. Identification of differences is important for designing mirtron prediction algorithms and may help to improve the understanding of mirtron functioning. So far, only simple, single-feature comparisons were reported. These are insensitive to complex feature relations. We quantified miRNAs with 25 features and showed that it is impossible to distinguish the two miRNA species using simple thresholds on any single feature. However, when using the Principal Component Analysis mirtrons and canonical miRNAs are grouped separately. Moreover, several methodologically diverse machine learning classifiers delivered high classification performance. Using feature selection algorithms we found features (e.g. bulges in the stem region), previously reported divergent in two classes, that did not contribute to improving classification accuracy, which suggests that they are not biologically meaningful. Finally, we proposed a combination of the most important features (including Guanine content, hairpin free energy and hairpin length) which convey a specific pattern, crucial for identifying mirtrons.
The observed variation in the purine composition of food after soaking in sake lees.[Pubmed:29750589]
Nucleosides Nucleotides Nucleic Acids. 2018;37(6):348-352.
In this study, we investigated the alterations in the purine composition of swordfish prepared using a traditional Japanese processing method of soaking in sake lees. These alterations are the byproducts of the yeast fermentation of rice-koji and are renowned for enhancing the umami nature of food. Using a conventional assay method for hydrolyzing all of the purines into four bases and our developed method for simultaneously analyzing purines, we observed the alterations in four purine bases in the soaked sake lees and swordfish. The findings showed that the total purine content, and hypoxanthine-related and Guanine-related purines in swordfish decreased after soaking in sake lees. We also analyzed the free purine composition and showed that the ratio of IMP in swordfish was decreased by soaking, while that of inosine in sake lees was increased by soaking swordfish in it.
Synthesis-identification integration: One-pot hydrothermal preparation of fluorescent nitrogen-doped carbon nanodots for differentiating nucleobases with the aid of multivariate chemometrics analysis.[Pubmed:29759232]
Talanta. 2018 Aug 1;185:491-498.
Most of the conventional multidimensional differential sensors currently need at least two-step fabrication, namely synthesis of probe(s) and identification of multiple analytes by mixing of analytes with probe(s), and were conducted using multiple sensing elements or several devices. In the study, we chose five different nucleobases (adenine, cytosine, Guanine, thymine, and uracil) as model analytes, and found that under hydrothermal conditions, sodium citrate could react directly with various nucleobases to yield different nitrogen-doped carbon nanodots (CDs). The CDs synthesized from different nucleobases exhibited different fluorescent properties, leading to their respective characteristic fluorescence spectra. Hence, we combined the fluorescence spectra of the CDs with advanced chemometrics like principle component analysis (PCA), hierarchical cluster analysis (HCA), K-nearest neighbor (KNN) and soft independent modeling of class analogy (SIMCA), to present a conceptually novel "synthesis-identification integration" strategy to construct a multidimensional differential sensor for nucleobase discrimination. Single-wavelength excitation fluorescence spectral data, single-wavelength emission fluorescence spectral data, and fluorescence Excitation-Emission Matrices (EEMs) of the CDs were respectively used as input data of the differential sensor. The results showed that the discrimination ability of the multidimensional differential sensor with EEM data set as input data was superior to those with single-wavelength excitation/emission fluorescence data set, suggesting that increasing the number of the data input could improve the discrimination power. Two supervised pattern recognition methods, namely KNN and SIMCA, correctly identified the five nucleobases with a classification accuracy of 100%. The proposed "synthesis-identification integration" strategy together with a multidimensional array of experimental data holds great promise in the construction of differential sensors.
NMR analyses on N-hydroxymethylated nucleobases - implications for formaldehyde toxicity and nucleic acid demethylases.[Pubmed:29767200]
Org Biomol Chem. 2018 May 30;16(21):4021-4032.
Formaldehyde is produced in cells by enzyme-catalysed demethylation reactions, including those occurring on N-methylated nucleic acids. Formaldehyde reacts with nucleobases to form N-hydroxymethylated adducts that may contribute to its toxicity/carcinogenicity when added exogenously, but the chemistry of these reactions has been incompletely defined. We report NMR studies on the reactions of formaldehyde with canonical/modified nucleobases. The results reveal that hydroxymethyl hemiaminals on endocyclic nitrogens, as observed with thymidine and uridine monophosphates, are faster to form than equivalent hemiaminals on exocyclic nitrogens; however, the exocyclic adducts, as formed with adenine, Guanine and cytosine, are more stable in solution. Nucleic acid demethylase (FTO)-catalysed hydroxylation of (6-methyl)adenosine results in (6-hydroxymethyl)adenosine as the major observed product; by contrast no evidence for a stable 3-hydroxymethyl adduct was accrued with FTO-catalysed oxidation of (3-methyl)thymidine. Collectively, our results imply N-hydroxymethyled adducts of nucleic acid bases, formed either by reactions with formaldehyde or via demethylase catalysis, have substantially different stabilities, with some being sufficiently stable to have functional roles in disease or the regulation of nucleic acid/nucleobase activity.
The influence of Cu(+) binding to hypoxanthine on stabilization of mismatches involving hypoxanthine and DNA bases: a DFT study.[Pubmed:29757083]
J Biomol Struct Dyn. 2019 May;37(8):1923-1934.
In the present work, the influence of Cu(+) binding to N3- and N7-positions of hypoxanthine on energetic, geometrical and topological properties of hypoxanthine-Guanine, hypoxanthine-adenine, hypoxanthine-cytosine, hypoxanthine-thymine and hypoxanthine-hypoxanthine mismatches is theoretically investigated. The calculations, in gas phase, are performed at B3LYP/6-311++G(3df,3pd) level of theory. Unlike the other mispairs, Cu(+) binding to N3-position of hypoxanthine causes the proton transfer process from enol form of hypoxanthine to imino forms of adenine and cytosine. This process also occurs in all mismatches having enol form of hypoxanthine when Cu(+) binds to N7-position of hypoxanthine. The mismatches are stabilized by hydrogen bonds. The influence of Cu(+) on hydrogen bonds is also examined by atoms in molecules (AIM) and natural bond orbital (NBO) analyses. Communicated by Ramaswamy H. Sarma.
Retinal adaptation to dim light vision in spectacled caimans (Caiman crocodilus fuscus): Analysis of retinal ultrastructure.[Pubmed:29753728]
Exp Eye Res. 2018 Aug;173:160-178.
It has been shown that mammalian retinal glial (Muller) cells act as living optical fibers that guide the light through the retinal tissue to the photoreceptor cells (Agte et al., 2011; Franze et al., 2007). However, for nonmammalian species it is unclear whether Muller cells also improve the transretinal light transmission. Furthermore, for nonmammalian species there is a lack of ultrastructural data of the retinal cells, which, in general, delivers fundamental information of the retinal function, i.e. the vision of the species. A detailed study of the cellular ultrastructure provides a basic approach of the research. Thus, the aim of the present study was to investigate the retina of the spectacled caimans at electron and light microscopical levels to describe the structural features. For electron microscopy, we used a superfast microwave fixation procedure in order to achieve more precise ultrastructural information than common fixation techniques. As result, our detailed ultrastructural study of all retinal parts shows structural features which strongly indicate that the caiman retina is adapted to dim light and night vision. Various structural characteristics of Muller cells suppose that the Muller cell may increase the light intensity along the path of light through the neuroretina and, thus, increase the sensitivity of the scotopic vision of spectacled caimans. Muller cells traverse the whole thickness of the neuroretina and thus may guide the light from the inner retinal surface to the photoreceptor cell perikarya and the Muller cell microvilli between the photoreceptor segments. Thick Muller cell trunks/processes traverse the layers which contain light-scattering structures, i.e., nerve fibers and synapses. Large Muller cell somata run through the inner nuclear layer and contain flattened, elongated Muller cell nuclei which are arranged along the light path and, thus, may reduce the loss of the light intensity along the retinal light path. The oblique arrangement of many Muller cell trunks/processes in the inner plexiform layer and the large Muller cell somata in the inner nuclear layer may suggest that light guidance through Muller cells increases the visual sensitivity. Furthermore, an adaptation of the caiman retina to low light levels is strongly supported by detailed ultrastructural data of other retinal parts, e.g. by (i) the presence of a Guanine-based retinal tapetum, (ii) the rod dominance of the retina, (iii) the presence of photoreceptor cell nuclei, which penetrate the outer limiting membrane, (iv) the relatively low densities of photoreceptor and neuronal cells which is compensated by (v) the presence of rods with long and thick outer segments, that may increase the probability of photon absorption. According to a cell number analysis, the central and temporal areas of the dorsal tapetal retina, which supports downward prey detection in darker water, are the sites of the highest diurnal contrast/color vision, i.e. cone vision and of the highest retinal light sensitivity, i.e. rod vision.


