Boehmeria nivea
Boehmeria nivea
1. The products in our compound library are selected from thousands of unique natural products; 2. It has the characteristics of diverse structure, diverse sources and wide coverage of activities; 3. Provide information on the activity of products from major journals, patents and research reports around the world, providing theoretical direction and research basis for further research and screening; 4. Free combination according to the type, source, target and disease of natural product; 5. The compound powder is placed in a covered tube and then discharged into a 10 x 10 cryostat; 6. Transport in ice pack or dry ice pack. Please store it at -20 °C as soon as possible after receiving the product, and use it as soon as possible after opening.
Natural products/compounds from Boehmeria nivea
- Cat.No. Product Name CAS Number COA
-
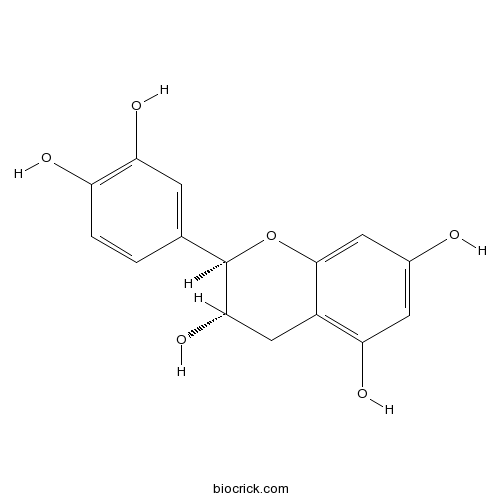
BCN1688
Catechin154-23-4
Instructions
-
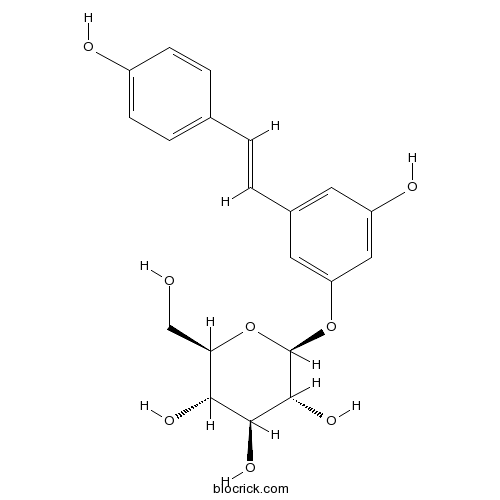
BCN5949
Polydatin27208-80-6
Instructions
-
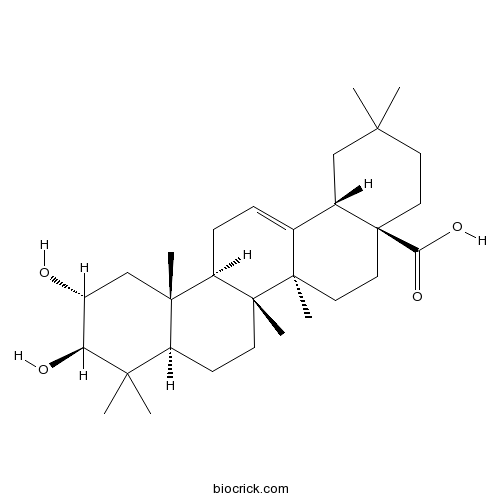
BCN5487
Crategolic acid4373-41-5
Instructions
-
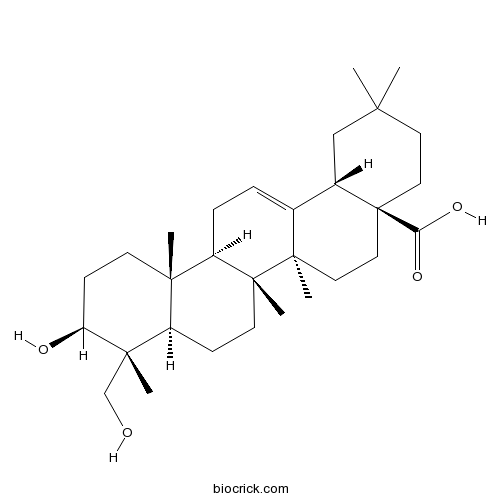
BCN5513
Hederagenin465-99-6
Instructions
-
BCN5649
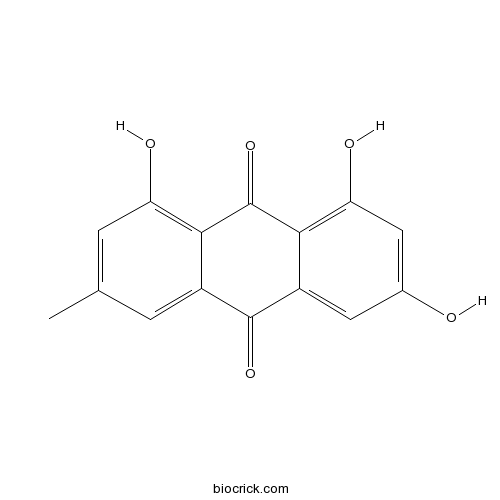
Emodin518-82-1
Instructions
Identification and expression characterization of the Phloem Protein 2 (PP2) genes in ramie (Boehmeria nivea L. Gaudich).[Pubmed: 30013165]
Phloem protein 2 (PP2) is one of the most abundant and enigmatic proteins in sieve elements and companion cells, which play important roles in the maintenance of morphology, photoassimilate transportation and wound protection in higher plants, but to date, no PP2 (BnPP2) genes had been identified in ramie. Here, a total of 15 full-length BnPP2 genes were identified. These BnPP2 genes exhibited different responses to abiotic stresses. Interestingly, the BnPP2 genes are more sensitive to insect pests than to other stresses. A study of the BnPP2-15 promoter revealed that pBnPP2-15 could drive specific GUS expression in the petiole, root and stamen and could also be induced by mechanical wounding and aphid infection in transgenic Arabidopsis lines. The subcellular localization of six BnPP2 proteins showed that GFP-BnPP2-1, GFP-BnPP2-6, GFP-BnPP2-7, GFP-BnPP2-9, GFP-BnPP2-11 and GFP-BnPP2-12 were predominantly located in the cytoplasm. These results provide useful information elucidating the functions of BnPP2 genes in ramie.
Potential use of high-throughput sequencing of soil microbial communities for estimating the adverse effects of continuous cropping on ramie (Boehmeria nivea L. Gaud).[Pubmed: 29750808]
Ramie (Boehmeria nivea L. Gaud) fiber, one of the most important natural fibers, is extracted from stem bark. Continuous cropping is the main obstacle to ramie stem growth and a major cause of reduced yields. Root-associated microbes play crucial roles in plant growth and health. In this study, we investigated differences between microbial communities in the soil of healthy and continuously cropped ramie plants, and sought to identify potential mechanisms whereby these communities could counteract the problems posed by continuous cropping. Paired-end Illumina MiSeq analysis of 16S rRNA and ITS gene amplicons was employed to study bacterial and fungal communities. Long-term monoculture of ramie significantly decreased fiber yields and altered soil microbial communities. Our findings revealed how microbial communities and functional diversity varied according to the planting year and plant health status. Soil bacterial diversity increased with the period of ramie monoculture, whereas no significant differences were observed for fungi. Sequence analyses revealed that Firmicutes, Proteobacteria, and Acidobacteria were the most abundant bacterial phyla. Firmicutes abundance decreased with the period of ramie monoculture and correlated positively with the stem length, stem diameter, and fiber yield. The Actinobacteria, Chloroflexi, and Zygomycota phyla exhibited a significant (P < 0.05) negative correlation with yields during continuous cultivation. Some Actinobacteria members showed reduced microbial diversity, which prevented continuous ramie cropping. Ascomycota, Zygomycota, and Basidiomycota were the main fungal phyla. The relatively high abundance of Bacillus observed in healthy ramie may contribute to disease suppression, thereby promoting ramie growth. In summary, soil weakness and increased disease in ramie plants after long-term continuous cropping can be attributed to changes in soil microbes, a reduction in beneficial microbes, and an accumulation of harmful microbes.
Draft genome sequence of ramie, Boehmeria nivea (L.) Gaudich.[Pubmed: 29423997]
Ramie, Boehmeria nivea (L.) Gaudich, family Urticaceae, is a plant native to eastern Asia, and one of the world's oldest fibre crops. It is also used as animal feed and for the phytoremediation of heavy metal-contaminated farmlands. Thus, the genome sequence of ramie was determined to explore the molecular basis of its fibre quality, protein content and phytoremediation. For further understanding ramie genome, different paired-end and mate-pair libraries were combined to generate 134.31 Gb of raw DNA sequences using the Illumina whole-genome shotgun sequencing approach. The highly heterozygous B. nivea genome was assembled using the Platanus Genome Assembler, which is an effective tool for the assembly of highly heterozygous genome sequences. The final length of the draft genome of this species was approximately 341.9 Mb (contig N50 = 22.62 kb, scaffold N50 = 1,126.36 kb). Based on ramie genome annotations, 30,237 protein-coding genes were predicted, and the repetitive element content was 46.3%. The completeness of the final assembly was evaluated by benchmarking universal single-copy orthologous genes (BUSCO); 90.5% of the 1,440 expected embryophytic genes were identified as complete, and 4.9% were identified as fragmented. Phylogenetic analysis based on single-copy gene families and one-to-one orthologous genes placed ramie with mulberry and cannabis, within the clade of urticalean rosids. Genome information of ramie will be a valuable resource for the conservation of endangered Boehmeria species and for future studies on the biogeography and characteristic evolution of members of Urticaceae.
Ramie (Boehmeria nivea)'s uranium bioconcentration and tolerance attributes.[Pubmed: 29395432]
The authors sampled and analyzed 15 species of dominant wild plants in Huanan uranium tailings pond in China, whose tailings' uranium contents were 3.21-120.52 μg/g. Among the 15 species of wild plants, ramie (Boehmeria nivea) had the strongest uranium bioconcentration and transfer capacities. In order to study the uranium bioconcentration and tolerance attributes of ramie in detail, and provide a reference for the screening remediation plants to phytoremedy on a large scale in uranium tailings pond, a ramie cultivar Xiangzhu No. 7 pot experiment was carried out. We found that both wild ramie and Xiangzhu No. 7 could bioconcentrate uranium, but there were two differences. One was wild ramie's shoots bioconcentrated uranium up to 20 μg/g (which can be regarded as the critical content value of the shoot of uranium hyperaccumulator) even the soil uranium content was as low as 5.874 μg/g while Xiangzhu No. 7's shoots could reach 20 μg/g only when the uranium treatment concentrations were 275 μg/g or more; the other was that all the transfer factors of 3 wild samples were >1, and the transfer factors of 27 out of 28 pot experiment samples were <1. Probably wild ramie was a uranium hyperaccumulator. Xiangzhu No. 7 satisfied the needs of uranium hyperaccumulator on accumulation capability, tolerance capability, bioconcentration factor, but not transfer capability, so Xiangzhu No. 7 was not a uranium hyperaccumulator. We analyzed the possible reasons why there were differences in the uranium bioconcentration and transfer attributes between wild ramie and Xiangzhu No. 7., and proposed the direction for further research. In our opinion, both the plants which bioconcentrate contaminants in the shoots and roots can act as phytoextractors. Although Xiangzhu No. 7's biomass and accumulation of uranium were concentrated on the roots, the roots were small in volume and easy to harvest. And Xiangzhu No. 7's cultivating skills and protection measures had been developed very well. Xiangzhu No. 7's whole bioconcentration factors and the roots' bioconcentration factors, which were 1.200-1.834 and 1.460-2.341, respectively, increased with the increases of uranium contents of pot soil when the soil's uranium contents are 25-175 μg/g, so it can act as a potential phytoextractor when Huanan uranium tailings pond is phytoremediated.
Draft genome analysis provides insights into the fiber yield, crude protein biosynthesis, and vegetative growth of domesticated ramie (Boehmeria nivea L. Gaud).[Pubmed: 29149285]
Plentiful bast fiber, a high crude protein content, and vigorous vegetative growth make ramie a popular fiber and forage crop. Here, we report the draft genome of ramie, along with a genomic comparison and evolutionary analysis. The draft genome contained a sequence of approximately 335.6 Mb with 42,463 predicted genes. A high-density genetic map with 4,338 single nucleotide polymorphisms (SNPs) was developed and used to anchor the genome sequence, thus, creating an integrated genetic and physical map containing a 58.2-Mb genome sequence and 4,304 molecular markers. A genomic comparison identified 1,075 unique gene families in ramie, containing 4,082 genes. Among these unique genes, five were cellulose synthase genes that were specifically expressed in stem bark, and 3 encoded a WAT1-related protein, suggesting that they are probably related to high bast fiber yield. An evolutionary analysis detected 106 positively selected genes, 22 of which were related to nitrogen metabolism, indicating that they are probably responsible for the crude protein content and vegetative growth of domesticated varieties. This study is the first to characterize the genome and develop a high-density genetic map of ramie and provides a basis for the genetic and molecular study of this crop.
Molecular Cloning, Recombinant Expression and Antifungal Activity of BnCPI, a Cystatin in Ramie (Boehmeria nivea L.).[Pubmed: 29019965]
None
Enhanced adsorption of hexavalent chromium by a biochar derived from ramie biomass (Boehmeria nivea (L.) Gaud.) modified with β-cyclodextrin/poly(L-glutamic acid).[Pubmed: 28852975]
This paper explored biochar modification to enhance biochar's ability to adsorb hexavalent chromium from aqueous solution. The ramie stem biomass was pyrolyzed and then treated by β-cyclodextrin/poly(L-glutamic acid) which contained plentiful functional groups. The pristine and modified biochar were characterized by FTIR, X-ray photoelectron spectroscopy, specific surface area, and zeta potential measurement. Results indicated that the β-cyclodextrin/poly(L-glutamic acid) was successfully bound to the biochar surface. Batch experiments were conducted to investigate the kinetics, isotherm, thermodynamics, and adsorption/desorption of Cr(VI). Adsorption capacities of CGA-biochar were significantly higher than that of the untreated biochar, and its maximum adsorption capacity could reach up to 197.21 mg/g at pH 2.0. Results also illustrated that sorption performance depended on initial solution pH; in addition, acidic condition was beneficial to the Cr(VI) uptake. Furthermore, the Cr(VI) uptake was significantly affected by the ion strength and cation species. This study demonstrated that CGA-biochar could be a potential adsorbent for Cr(VI) pollution control.


