Suramin hexasodium saltNon-selective P2 antagonist CAS# 129-46-4 |
- Vandetanib (ZD6474)
Catalog No.:BCC3883
CAS No.:443913-73-3
- Brivanib Alaninate (BMS-582664)
Catalog No.:BCC1240
CAS No.:649735-63-7
- Crenolanib (CP-868596)
Catalog No.:BCC3671
CAS No.:670220-88-9
- Apatinib
Catalog No.:BCC5099
CAS No.:811803-05-1
- Flumatinib mesylate
Catalog No.:BCC3970
CAS No.:895519-91-2
Quality Control & MSDS
Number of papers citing our products
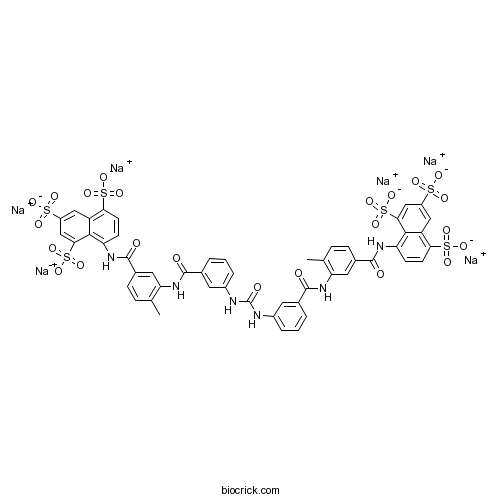
Chemical structure

3D structure
| Cas No. | 129-46-4 | SDF | Download SDF |
| PubChem ID | 8514 | Appearance | Powder |
| Formula | C51H34N6Na6O23S6 | M.Wt | 1429.15 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Synonyms | Suramin hexasodium salt; BAY-205; NF-060 | ||
| Solubility | H2O : ≥ 200 mg/mL (139.94 mM) DMSO : 5.6 mg/mL (3.92 mM; Need warming) *"≥" means soluble, but saturation unknown. | ||
| Chemical Name | hexasodium;8-[[4-methyl-3-[[3-[[3-[[2-methyl-5-[(4,6,8-trisulfonatonaphthalen-1-yl)carbamoyl]phenyl]carbamoyl]phenyl]carbamoylamino]benzoyl]amino]benzoyl]amino]naphthalene-1,3,5-trisulfonate | ||
| SMILES | CC1=C(C=C(C=C1)C(=O)NC2=C3C(=CC(=CC3=C(C=C2)S(=O)(=O)[O-])S(=O)(=O)[O-])S(=O)(=O)[O-])NC(=O)C4=CC(=CC=C4)NC(=O)NC5=CC=CC(=C5)C(=O)NC6=C(C=CC(=C6)C(=O)NC7=C8C(=CC(=CC8=C(C=C7)S(=O)(=O)[O-])S(=O)(=O)[O-])S(=O)(=O)[O-])C.[Na+].[Na+].[Na+].[Na+].[Na+].[Na+] | ||
| Standard InChIKey | VAPNKLKDKUDFHK-UHFFFAOYSA-H | ||
| Standard InChI | InChI=1S/C51H40N6O23S6.6Na/c1-25-9-11-29(49(60)54-37-13-15-41(83(69,70)71)35-21-33(81(63,64)65)23-43(45(35)37)85(75,76)77)19-39(25)56-47(58)27-5-3-7-31(17-27)52-51(62)53-32-8-4-6-28(18-32)48(59)57-40-20-30(12-10-26(40)2)50(61)55-38-14-16-42(84(72,73)74)36-22-34(82(66,67)68)24-44(46(36)38)86(78,79)80;;;;;;/h3-24H,1-2H3,(H,54,60)(H,55,61)(H,56,58)(H,57,59)(H2,52,53,62)(H,63,64,65)(H,66,67,68)(H,69,70,71)(H,72,73,74)(H,75,76,77)(H,78,79,80);;;;;;/q;6*+1/p-6 | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||
| Description | Non-selective P2 purinergic antagonist. Also blocks calmodulin binding to recognition sites and G protein coupling to G protein-coupled receptors. Increases open probability of ryanodine receptor (RyR) channels. Also acts as a competitive α1β2γ2 GABAA receptor antagonist. Anticancer and antiviral agent. |

Suramin hexasodium salt Dilution Calculator

Suramin hexasodium salt Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 0.6997 mL | 3.4986 mL | 6.9972 mL | 13.9943 mL | 17.4929 mL |
| 5 mM | 0.1399 mL | 0.6997 mL | 1.3994 mL | 2.7989 mL | 3.4986 mL |
| 10 mM | 0.07 mL | 0.3499 mL | 0.6997 mL | 1.3994 mL | 1.7493 mL |
| 50 mM | 0.014 mL | 0.07 mL | 0.1399 mL | 0.2799 mL | 0.3499 mL |
| 100 mM | 0.007 mL | 0.035 mL | 0.07 mL | 0.1399 mL | 0.1749 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
Suramin sodium salt is a polysulfonated naphthylurea with various biological activities. Suramin sodium salt is a DNA topoisomerase II inhibitor with an IC50 of 5 μM.
In Vitro:Suramin inhibits cell proliferation and DNA synthesis in cultured HeLa cells. The replication of SV40 DNA is completely abolished by 40 μM suramin. DNA polymerase α is sensitive to lower concentrations of suramin (IC50=8 µM) than is DNA polymerase δ (IC50=36 µM), whereas DNA polymerase β is relatively insensitive to the drug (IC50 of 90 µM)[1]. Suramin is a potent inhibitor of DNA strand exchange and ATPase activities of bacterial RecA proteins. Suramin inhibits RecA-catalysed proteolytic cleavage of the LexA repressor. The mechanism underlying such inhibitory actions of suramin involves its ability to disassemble RecA–single-stranded DNA filaments[2]. Suramin is a potent inhibitor of the nuclear enzyme DNA topoisomerase II. Suramin inhibits purified yeast topoisomerase II with an IC50 of about 5 μM[3].
In Vivo:Treatment with suramin shows lower values for pulmonary artery pressure, right ventricular hypertrophy, and distal vessel muscularization on day 21 compared to control rats. Suramin treatment suppresses PA-SMC proliferation and attenuates both the inflammatory response and the deposition of collagen[4].
References:
[1]. Jindal HK, et al. Suramin affects DNA synthesis in HeLa cells by inhibition of DNA polymerases. Cancer Res. 1990 Dec 15;50(24):7754-7.
[2]. Nautiyal A, et al. Suramin is a potent and selective inhibitor of Mycobacterium tuberculosis RecA protein and the SOS response: RecA as a potential target for antibacterial drug discovery. J Antimicrob Chemother. 2014 Jul;69(7):1834-43.
[3]. Bojanowski K, et al. Suramin is an inhibitor of DNA topoisomerase II in vitro and in Chinese hamster fibrosarcomacells. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3025-9.
[4]. Izikki M, et al. The beneficial effect of suramin on monocrotaline-induced pulmonary hypertension in rats. PLoS One. 2013 Oct 15;8(10):e77073.
- Maohuoside A
Catalog No.:BCN5348
CAS No.:128988-55-6
- Fargesol
Catalog No.:BCN6421
CAS No.:128855-64-1
- Fmoc-D-Asp(OtBu)-OH
Catalog No.:BCC3471
CAS No.:12883-39-3
- Mycophenolate Mofetil
Catalog No.:BCC2290
CAS No.:128794-94-5
- Kalopanaxsaponin H
Catalog No.:BCN2572
CAS No.:128730-82-5
- Eucamalduside A
Catalog No.:BCN7321
CAS No.:1287220-29-4
- FRAX597
Catalog No.:BCC4172
CAS No.:1286739-19-2
- 1,6-O,O-Diacetylbritannilactone
Catalog No.:BCN7792
CAS No.:1286694-67-4
- PyBOP
Catalog No.:BCC2820
CAS No.:128625-52-5
- Z(2-Br)-Osu
Catalog No.:BCC2806
CAS No.:128611-93-8
- Ospemifene
Catalog No.:BCC5557
CAS No.:128607-22-7
- Pingpeimine C
Catalog No.:BCN8411
CAS No.:128585-96-6
- Methysergide maleate
Catalog No.:BCC5698
CAS No.:129-49-7
- SP 600125
Catalog No.:BCC2474
CAS No.:129-56-6
- Buclizine HCl
Catalog No.:BCC4516
CAS No.:129-74-8
- Rivastigmine Tartrate
Catalog No.:BCC3851
CAS No.:129101-54-8
- Evodosin A
Catalog No.:BCN7322
CAS No.:1291053-38-7
- ENMD-2076 L-(+)-Tartaric acid
Catalog No.:BCC2185
CAS No.:1291074-87-7
- CGRP 8-37 (rat)
Catalog No.:BCC5717
CAS No.:129121-73-9
- 2-(2,2-Dimethyl-1,3-dioxolan-4-yl)propane-1,2-diol
Catalog No.:BCC8475
CAS No.:129141-48-6
- Gancaonin M
Catalog No.:BCN4757
CAS No.:129145-51-3
- Biotin-HPDP
Catalog No.:BCC3583
CAS No.:129179-83-5
- Iberiotoxin
Catalog No.:BCC6932
CAS No.:129203-60-7
- (2S,3S)-(-)-Glucodistylin
Catalog No.:BCN6156
CAS No.:129212-92-6
Functional regulation of the cardiac ryanodine receptor by suramin and calmodulin involves multiple binding sites.[Pubmed:15102954]
Mol Pharmacol. 2004 May;65(5):1258-68.
Suramin and structurally related compounds increase not only the open probability (P(o)) of ryanodine receptor (RyR) channels but also the single-channel conductance in a unique characteristic manner. In this report, we examine the mechanisms underlying the complex changes to cardiac RyR channel function caused by suramin and the evidence that these changes result from an interaction with calmodulin (CaM) binding sites. In the presence of 100 microM cytosolic Ca(2+), we demonstrate that suramin exerts a triphasic effect on P(o), indicating the presence of high-, intermediate-, and low-affinity suramin binding sites. The effects of suramin binding to high-affinity sites are Ca(2+)-dependent; P(o) is decreased and seems to result from a reduction in the sensitivity of the channel to cytosolic Ca(2+). We suggest that this site is the CaM inhibition site. Suramin also binds to intermediate-affinity sites that mediate an increase in P(o) and an increase in conductance. Cytosolic Ca(2+) is not an absolute requirement for the effects mediated via intermediate-affinity suramin sites. The suramin-induced increase in P(o) and conductance are both concentration-dependent. The correlation between the increase in P(o) and increase in conductance indicates that the binding events which produce an increase in P(o) also lead to an increase in conductance and, because the effect is concentration-dependent, multiple suramin molecules must bind to produce the maximum effect. The low-affinity suramin binding sites are inhibition sites and mediate a reduction in P(o) caused by changes to both open and closed lifetimes.
Suramin and the suramin analogue NF307 discriminate among calmodulin-binding sites.[Pubmed:11311147]
Biochem J. 2001 May 1;355(Pt 3):827-33.
Calmodulin-binding sites on target proteins show considerable variation in primary sequence; hence compounds that block the access of calmodulin to these binding sites may be more selective than compounds that inactivate calmodulin. Suramin and its analogue NF307 inhibit the interaction of calmodulin with the ryanodine receptor. We have investigated whether inhibition of calmodulin binding to target proteins is a general property of these compounds. Suramin inhibited binding of [(125)I]calmodulin to porcine brain membranes and to sarcoplasmic reticulum from skeletal muscle (IC(50)=4.9+/-1.2 microM and 19.9+/-1.8 microM, respectively) and blocked the cross-linking of [(125)I]calmodulin to some, but not all, target proteins in brain membranes by [(125)I]calmodulin. Four calmodulin-binding proteins were purified [ryanodine receptor-1 (RyR1) from rabbit skeletal muscle, neuronal NO synthase (nNOS) from Sf9 cells, G-protein betagamma dimers (Gbetagamma) from porcine brain and a glutathione S-transferase-fusion protein comprising the C-terminal calmodulin-binding domain of the metabotropic glutamate receptor 7A (GST-CmGluR7A) from bacterial lysates]. Three of the proteins employed (Gbetagamma, GST-CmGluR7A and RyR1) display a comparable affinity for calmodulin (in the range of 50-70 nM). Nevertheless, suramin and NF307 only blocked the binding of Gbetagamma and RyR1 to calmodulin-Sepharose. In contrast, the association of GST-CmGluR7A and nNOS was not impaired, whereas excess calmodulin uniformly displaced all proteins from the matrix. Thus suramin and NF307 are prototypes of a new class of calmodulin antagonists that do not interact directly with calmodulin but with calmodulin-recognition sites. In addition, these compounds discriminate among calmodulin-binding motifs.
Inhibition of receptor/G protein coupling by suramin analogues.[Pubmed:8700151]
Mol Pharmacol. 1996 Aug;50(2):415-23.
Suramin analogues act as direct antagonists of heterotrimeric G proteins because they block the rate-limiting step of G protein activation (i.e., the dissociation of GDP prebound to the G protein alpha subunit). We have used the human brain A1 adenosine receptor and the rat striatal D2 dopamine receptor, two prototypical Gi/G(o)-coupled receptors, as a model system to test whether the following analogues suppress the receptor-dependent activation of G proteins: 8-(3-nitrobenzamido)-1,3,5-naphthalenetrisulfonic acid (NF007), 8-(3-(3-nitrobenzamido)-benzamido)-1,3,5-naphthalenetrisulfonic acid (NF018); 8,8'-(carbonylbis(imino-3,1-phenylene))bis-(1,3,5-naphthalenetr isulfonic acid) (NF023); 8,8'-(carbonylbis(imino-3,1-phenylene)carbonylimino-(3,1- phenylene)) bis(1,3,5-naphthalenetrisulfonic acid) (NF037); and suramin. Suramin and its analogues inhibit the formation of the agonist-specific ternary complex (agonist/receptor/G protein). This inhibition is (i) quasicompetitive with respect to agonist binding in that it can be overcome by increasing receptor occupancy but (ii) does not result from an interaction of the analogues with the ligand binding pocket of the receptors because the binding of antagonists or of agonists in the absence of functional receptor/G protein interaction is not affected. In addition to suppressing the spontaneous release of GDP from defined G protein alpha subunits, suramin and its analogues reduce receptor-catalyzed guanine nucleotide exchange. The site, to which suramin analogues bind, overlaps with the docking site for the receptor on the G protein alpha subunit. The structure-activity relationships for inhibition of agonist binding to the A1 adenosine receptor (suramin > NF037 > NF023) and of agonist binding to the inhibition D2 dopamine receptor (suramin = NF037 > NF023 > NF018) differ. Thus, NF037 discriminates between the ternary complexes formed by the agonist-liganded D2 dopamine receptors and those formed by the A1 adenosine receptor with > 10-fold selectivity. Therefore, our results also show that inhibitors can be identified that selectively uncouple specific receptor/G protein tandems.
PPADS and suramin as antagonists at cloned P2Y- and P2U-purinoceptors.[Pubmed:8762097]
Br J Pharmacol. 1996 Jun;118(3):704-10.
1. The effect of suramin and pyridoxalphosphate-6-azophenyl-2',4'-disulphonic acid (PPADS) on the stimulation of phospholipase C in 1321N1 cells transfected with the human P2U-purinoceptor (h-P2U-1321N1 cells) or with the turkey P2Y-purinoceptor (t-P2Y-1321N1 cells) was investigated. 2-Methylthioadenosine triphosphate (2MeSATP) was used as the agonist at t-P2Y-1321N1 cells and uridine triphosphate (UTP) at h-P2U-1321N1 cells. 2. Suramin caused a parallel shift to the right of the concentration-response curves for 2MeSATP in the t-P2Y-1321N1 cells, yielding a Schild plot with a slope of 1.16 +/- 0.08 and a pA2 value of 5.77 +/- 0.11. 3. Suramin also caused a shift to the right of concentration-response curves for UTP in the h-P2U-1321N1 cells, and on Schild plots gave a slope different from unity (1.57 +/- 0.19) and an apparent pA2 value of 4.32 +/- 0.13. Suramin was therefore a less potent antagonist at the P2U-purinoceptor than the P2Y-purinoceptor. 4. In the presence of the ectonucleotidase inhibitor, ARL 67156 (6-N,N-diethyl-beta,gamma-dibromomethylene-D-ATP) there was no significant difference in the EC50 or shapes of curves with either cell type, and no difference in pA2 values for suramin. 5. PPADS caused an increase in the EC50 for 2MeSATP in the t-P2Y-1321N1 cells. The Schild plot had a slope different from unity (0.55 +/- 0.15) and an X-intercept corresponding to an apparent pA2 of 5.98 +/- 0.65. 6. PPADS up to 30 microM had no effect on the concentration-response curve for UTP with the h-P2U-1321N1 cells. 7. In conclusion, suramin and PPADS show clear differences in their action at the 2 receptor types, in each case being substantially more effective as an antagonist at the P2Y-purinoceptor than at the P2U-purinoceptor. Ectonucleotidase breakdown had little influence on the nature of the responses at the two receptor types, or in their differential sensitivity to suramin.


