Sieber LinkerCAS# 3722-51-8 |
- PHA-665752
Catalog No.:BCC1181
CAS No.:477575-56-7
- (R)-Crizotinib
Catalog No.:BCC1284
CAS No.:877399-52-5
- Tivantinib (ARQ 197)
Catalog No.:BCC3688
CAS No.:905854-02-6
Quality Control & MSDS
3D structure
Package In Stock
Number of papers citing our products
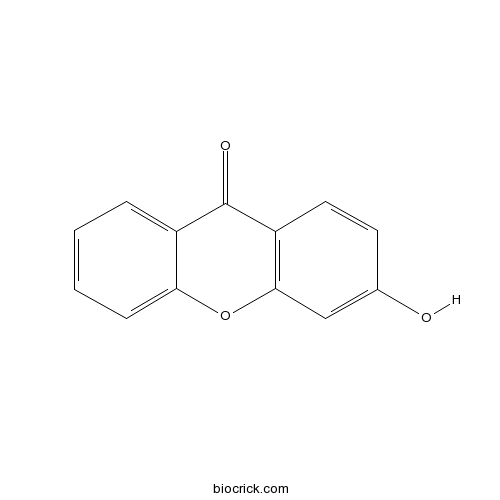
| Cas No. | 3722-51-8 | SDF | Download SDF |
| PubChem ID | 5376013 | Appearance | Powder |
| Formula | C13H8O3 | M.Wt | 212.2 |
| Type of Compound | N/A | Storage | Desiccate at -20°C |
| Solubility | Soluble in Chloroform,Dichloromethane,Ethyl Acetate,DMSO,Acetone,etc. | ||
| Chemical Name | 3-hydroxyxanthen-9-one | ||
| SMILES | C1=CC=C2C(=C1)C(=O)C3=C(O2)C=C(C=C3)O | ||
| Standard InChIKey | XCJHDJAODLKGLG-UHFFFAOYSA-N | ||
| Standard InChI | InChI=1S/C13H8O3/c14-8-5-6-10-12(7-8)16-11-4-2-1-3-9(11)13(10)15/h1-7,14H | ||
| General tips | For obtaining a higher solubility , please warm the tube at 37 ℃ and shake it in the ultrasonic bath for a while.Stock solution can be stored below -20℃ for several months. We recommend that you prepare and use the solution on the same day. However, if the test schedule requires, the stock solutions can be prepared in advance, and the stock solution must be sealed and stored below -20℃. In general, the stock solution can be kept for several months. Before use, we recommend that you leave the vial at room temperature for at least an hour before opening it. |
||
| About Packaging | 1. The packaging of the product may be reversed during transportation, cause the high purity compounds to adhere to the neck or cap of the vial.Take the vail out of its packaging and shake gently until the compounds fall to the bottom of the vial. 2. For liquid products, please centrifuge at 500xg to gather the liquid to the bottom of the vial. 3. Try to avoid loss or contamination during the experiment. |
||
| Shipping Condition | Packaging according to customer requirements(5mg, 10mg, 20mg and more). Ship via FedEx, DHL, UPS, EMS or other couriers with RT, or blue ice upon request. | ||

Sieber Linker Dilution Calculator

Sieber Linker Molarity Calculator
| 1 mg | 5 mg | 10 mg | 20 mg | 25 mg | |
| 1 mM | 4.7125 mL | 23.5627 mL | 47.1254 mL | 94.2507 mL | 117.8134 mL |
| 5 mM | 0.9425 mL | 4.7125 mL | 9.4251 mL | 18.8501 mL | 23.5627 mL |
| 10 mM | 0.4713 mL | 2.3563 mL | 4.7125 mL | 9.4251 mL | 11.7813 mL |
| 50 mM | 0.0943 mL | 0.4713 mL | 0.9425 mL | 1.885 mL | 2.3563 mL |
| 100 mM | 0.0471 mL | 0.2356 mL | 0.4713 mL | 0.9425 mL | 1.1781 mL |
| * Note: If you are in the process of experiment, it's necessary to make the dilution ratios of the samples. The dilution data above is only for reference. Normally, it's can get a better solubility within lower of Concentrations. | |||||

Calcutta University

University of Minnesota

University of Maryland School of Medicine

University of Illinois at Chicago

The Ohio State University

University of Zurich

Harvard University

Colorado State University

Auburn University

Yale University

Worcester Polytechnic Institute

Washington State University

Stanford University

University of Leipzig

Universidade da Beira Interior

The Institute of Cancer Research

Heidelberg University

University of Amsterdam

University of Auckland

TsingHua University

The University of Michigan

Miami University

DRURY University

Jilin University

Fudan University

Wuhan University

Sun Yat-sen University

Universite de Paris

Deemed University

Auckland University

The University of Tokyo

Korea University
Sieber Linker
- PIK-75
Catalog No.:BCC1163
CAS No.:372196-77-5
- Citromycin
Catalog No.:BCN7459
CAS No.:37209-30-6
- Capsidiol
Catalog No.:BCC8140
CAS No.:37208-05-2
- L-Citruline
Catalog No.:BCN2692
CAS No.:372-75-8
- SB 612111 hydrochloride
Catalog No.:BCC7714
CAS No.:371980-94-8
- ZAPA sulfate
Catalog No.:BCC6563
CAS No.:371962-01-5
- YM201636
Catalog No.:BCC4996
CAS No.:371942-69-7
- PI-103 Hydrochloride
Catalog No.:BCC1860
CAS No.:371935-79-4
- PI-103
Catalog No.:BCC1162
CAS No.:371935-74-9
- Flavoxate hydrochloride
Catalog No.:BCC5208
CAS No.:3717-88-2
- 4'-Amino-3',5'-dichloroacetophenone
Catalog No.:BCC8678
CAS No.:37148-48-4
- Murrangatin
Catalog No.:BCN5426
CAS No.:37126-91-3
- Wilforgine
Catalog No.:BCN5427
CAS No.:37239-47-7
- Wilfortrine
Catalog No.:BCN3085
CAS No.:37239-48-8
- Wilfordine
Catalog No.:BCN3083
CAS No.:37239-51-3
- TCS OX2 29
Catalog No.:BCC7670
CAS No.:372523-75-6
- Sennoside C
Catalog No.:BCN1004
CAS No.:37271-16-2
- Sennoside D
Catalog No.:BCN1005
CAS No.:37271-17-3
- H-D-Tyr-OMe.HCl
Catalog No.:BCC3135
CAS No.:3728-20-9
- Flavokawain C
Catalog No.:BCN8456
CAS No.:37308-75-1
- 3-Quinuclidinone
Catalog No.:BCC8642
CAS No.:3731-38-2
- Decloxizine
Catalog No.:BCC5529
CAS No.:3733-63-9
- Cephaeline Hydrochloride
Catalog No.:BCC8307
CAS No.:3738-70-3
- DS2
Catalog No.:BCC7748
CAS No.:374084-31-8
The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane.[Pubmed:16443657]
Biophys J. 2006 Apr 15;90(8):2843-51.
In the plasma membrane, syntaxin 1 and syntaxin 4 clusters define sites at which secretory granules and caveolae fuse, respectively. It is widely believed that lipid phases are mandatory for cluster formation, as cluster integrity depends on cholesterol. Here we report that the native lipid environment is not sufficient for correct syntaxin 1 clustering and that additional cytoplasmic protein-protein interactions, primarily involving the SNARE motif, are required. Apparently no specific cofactors are needed because i), clusters form equally well in nonneuronal cells, and ii), as revealed by nanoscale subdiffraction resolution provided by STED microscopy, the number of clusters directly depends on the syntaxin 1 concentration. For syntaxin 4 clustering the N-terminal domain and the linker region are also dispensable. Moreover, clustering is specific because in both cluster types syntaxins mutually exclude one another at endogenous levels. We suggest that the SNARE motifs of syntaxin 1 and 4 mediate specific syntaxin clustering by homooligomerization, thereby spatially separating sites for different biological activities. Thus, syntaxin clustering represents a mechanism of membrane patterning that is based on protein-protein interactions.
New heterocyclic beta-sheet ligands with peptidic recognition elements.[Pubmed:15287758]
J Org Chem. 2004 Aug 6;69(16):5168-78.
A detailed and comprehensive overview is presented about the design, modeling, and synthesis, as well as spectroscopic characterization, of a new class of beta-sheet ligands. The characteristic feature of these compounds is a peptidic chimeric structure formed from a specific combination of aminopyrazolecarboxylic acids with naturally occurring alpha-amino acids. These hybrid peptides are designed with the aid of molecular modeling to exist mainly in an extended conformation. All their hydrogen bond donors and acceptors can be aligned at the bottom face in such a way that a perfect complementarity toward beta-sheets is obtained. Thus the aminopyrazoles impart rigidity and a highly efficient DAD sequence for the recognition of whole dipeptide fragments, whereas the natural alpha-amino acids are designed to mimick recognition sites in proteins, ultimately leading to sequence-selective protein recognition. The synthetic protocols either rely upon solution phase peptide coupling with a PMB protecting group strategy or solid-phase peptide coupling based on the Fmoc strategy, using the same protecting group. In solution, a key building block was prepared by catalytic reduction of a nitropyrazolecarboxylic acid precursor. Subsequently, it was (N-1)-protected with a PMB group, and elongated by HCTU- or T3P-assisted peptide coupling with dipeptide fragments, followed by PyClop-assisted coupling with another nitropyrazolecarboxylic acid building block. Final simultaneous deprotection of all PMB groups with hot TFA completed the high-yield protocol, which works racemization-free. After preparing a similar key building block with an Fmoc protection at N-3, we developed a strategy suitable for automated synthesis of larger hybrid ligands on a peptide synthesizer. Attachment of the first amino acid to a polystyrene resin over the Sieber amide linker is followed by an iterative sequence consisting of Fmoc deprotection with piperidine and subsequent coupling with natural alpha-amino acid via HATU/HOAt. High yields of free hybrid peptides are obtained after mild acidic cleavage from the resin, followed by deprotection of the PMB groups with hot TFA. The new aminopyrazole peptide hybrid compounds were characterized by various spectroscopic measurements including CD spectra, VT, and ROESY NMR experiments. All these accumulated data indicate the absence of any intramolecular hydrogen bonds and strongly support an extended conformation in solution, ideal for docking on to solvent-exposed beta-sheets in proteins. Initial results from aggregation tests of pathological proteins with these and related ligands look extremely promising.
A photolabile linker for the mild and selective cleavage of enriched biomolecules from solid support.[Pubmed:19821566]
J Org Chem. 2009 Nov 6;74(21):8476-9.
Selective release of enriched biomolecules from solid support is a desirable goal in proteomic and metabolomic studies. Here we demonstrate that photocleavage of a light-sensitive phenacyl ester bond is a suitable alternative cleavage strategy for the selective release of enriched biomolecules form avidin beads circumventing the disadvantages of conventional heat denaturation procedures.
Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.[Pubmed:23175611]
Nucleic Acids Res. 2013 Jan;41(2):1343-54.
Recognition of the 3'-splice site is a key step in pre-mRNA splicing and accomplished by a dynamic complex comprising splicing factor 1 (SF1) and the U2 snRNP auxiliary factor 65-kDa subunit (U2AF65). Both proteins mediate protein-protein and protein-RNA interactions for cooperative RNA-binding during spliceosome assembly. Here, we report the solution structure of a novel helix-hairpin domain in the N-terminal region of SF1 (SF1(NTD)). The nuclear magnetic resonance- and small-angle X-ray scattering-derived structure of a complex of the SF1(NTD) with the C-terminal U2AF homology motif domain of U2AF65 (U2AF65(UHM)) reveals that, in addition to the known U2AF65(UHM)-SF1 interaction, the helix-hairpin domain forms a secondary, hydrophobic interface with U2AF65(UHM), which locks the orientation of the two subunits. Mutational analysis shows that the helix hairpin is essential for cooperative formation of the ternary SF1-U2AF65-RNA complex. We further show that tandem serine phosphorylation of a conserved Ser80-Pro81-Ser82-Pro83 motif rigidifies a long unstructured linker in the SF1 helix hairpin. Phosphorylation does not significantly alter the overall conformations of SF1, SF1-U2AF65 or the SF1-U2AF65-RNA complexes, but slightly enhances RNA binding. Our results indicate that the helix-hairpin domain of SF1 is required for cooperative 3'-splice site recognition presumably by stabilizing a unique quaternary arrangement of the SF1-U2AF65-RNA complex.
An improved aldehyde linker for the solid phase synthesis of hindered amides.[Pubmed:16468778]
J Org Chem. 2006 Feb 17;71(4):1322-9.
A novel aldehyde dual-linker system has been developed for the solid phase synthesis of sterically hindered amides. The linker [5-(4-formyl-3-hydroxyphenoxy)pentanoic acid] exploits an intramolecular oxygen-nitrogen acyl transfer mechanism to prepare compounds that are unattainable with current commercially available linkers. A dual linker system, exploiting the hyper-acid labile Sieber amide linker as part of the construct, enabled the initial reductive alkylation reactions of hindered amines and their subsequent acylation with a range of carboxylic acids with varying stereoelectronic properties to be monitored. Simple acylation conditions (HBTU/HOBt/NMM) sufficed to provide near quantitative reaction of test acids with support-bound hindered amines, reaction conditions which failed when commercial linkers were used.


